Seldonian ML
GitHub
Tutorial N: Using multiple datasets with the toolkit
Contents
Introduction
In all of the previous tutorials, we have assumed that there is a single dataset $D$ for a given Seldonian machine learning problem. However, this assumption was made for simplicity, and it can be dropped without violating the high-confidence guarantees of Seldonian algorithms. In this tutorial, we refer to the dataset that is used to evaluate the primary objective during candidate selection as the primary dataset and the datasets that are used to evalute the constraints (both during candidate selection and the safety test) as additional datasets. A common scenario where this may arise is when one has a small fraction of data with labels for the sensitive attributes (e.g., age, race, gender) and a much larger fraction without these labels. The sensitive attribute labels are often only necessary for evaluating the behavioral constraints and unnecessary for evaluating a primary objective function (e.g., mean squared error, cross entropy).
In this tutorial, we introduce a feature for specifying additional datasets. We also introduce a related feature that enables specifying candidate and safety datasets directly, as opposed to having the toolkit split the provided dataset using the frac_data_in_safety parameter. We will use a modified version of the dataset described in Tutorial E: Predicting student GPAs from application materials with fairness guarantees to demonstrate these features. We highly recommend acquainting yourself with Tutorial E before proceeding here.
Briefly, the dataset used in Tutorial E consists of 43,303 data points, where each point contains test scores on 9 entrance exams for a single student. The (binary) gender of all 43,303 students is known. In this tutorial, we invent a scenario where the gender of only 30% of the students is known. The five behavioral constraints studied in Tutorial E were disparate impact, demographic parity, equalized odds, equal opportunity and predictive equality. Each of these constraints used the gender attribute in some capacity. For example, the demographic parity constraint ensures that the positive rates (probability of predicting a GPA > 3.0) for male and female applicants are within some small fraction of each other, with probability $1-\delta$.
In the new scenario described above, each fairness constraint can only be evaluated using 30% of the data because it relies on the gender attribute. However, the primary objective function for this problem, the logistic loss (see Tutorial E), does not use the gender attribute. As a result, we can use 100% of the data when evaluating the primary objective.
Note 1: It is important to ensure that if there are shared data points between the primary dataset and additional datasets that no data points that are used in candidate selection from any dataset (whether primary or additional) are used during the safety test. We leave this up to the user; there are no checks in the Seldonian Toolkit to ensure that no data are shared.
Note 2: Additional datasets are currently only supported for supervised learning problems within the toolkit.
Outline
In this tutorial, you will learn how to:
- Specify additional datasets for evaluating behavioral constraints vs. the primary objective function.
- Provide candidate and safety datasets as direct inputs (for both the primary dataset and additional datasets).
Dataset preparation
The scenario in this tutorial uses two datasets: i) a primary dataset which consists of the original features and GPA class labels, but no gender attributes, ii) an additional dataset containing 30% of the same data but with gender attributes included. This is an invented scenario intended to mimic a realistic scenario where only a fraction of the total amount of data contain sensitive attribute labels. We created a Jupyter notebook implementing the steps for creating these datasets. The notebook also demonstrates how to create candidate and safety splits in a way to ensure that no data is shared between any candidate datasets and any safety datasets (whether primary or additional). The resulting data files and metadata files that are produced by this notebook can be found here: https://github.com/seldonian-toolkit/Engine/tree/main/examples/GPA_addl_dataset/.
The Seldonian ML problem we are addressing is similar to the one addressed in Tutorial E. In that tutorial, we considered five different definitions of fairness to apply to the problem of predicting whether students would have high or low GPAs based on nine entrance examination scores. For simplicity, we are going to focus on a single constraint in this tutorial: demographic parity. The constraint string we will use for this tutorial is the same as the one used in Tutorial E:
$$
``\text{abs((PR | [M]) - (PR | [F])) <= 0.2"}
\label{dpstr}
$$
As in Tutorial E, we are going to apply this constraint with $\delta=0.05$.
Creating the specification object
To run the Seldonian algorithm using the toolkit, we need to create a specification (or "spec") object. This is where we will supply our additional dataset to be evaluated on the constraint. In versions $>=0.8.8$ of the Seldonian Engine, there is a new parameter additional_datasets in the Spec class (and all child classes). This parameter expects a nested dictionary that maps the constraint strings to dictionaries that map that constraint's unique base node strings to the additional datasets one wants to use to bound those base nodes. Below are the rules of this dictionary:
-
The outermost key is the constraint string for a given parse tree. Its value is itself a dictionary, whose keys are the unique base node strings, as they appear in the constraint string, and whose values are seldonian.dataset.DataSet objects (or any child class objects of DataSet).
-
If a constraint string is missing from the dictionary, each base node in the corresponding tree will use the primary dataset by default. Note that this can lead to an error if the constraint requires access to a sensitive attribute that is not present in the primary dataset.
-
If a base node string is missing from a constraint's dictionary, then that base node will be evaluated using the primary dataset by default. Again, note that this can lead to errors.
-
The constraint string keys to the
additional_datasets dictionary need to match the constraint_str attribute of the built parse trees exactly. Note that the constraint string that one supplies when building the parse tree may differ from the constraint_str attribute of the built tree. When parse trees are built, they may rearrange the constraint string so that it is in an internally consistent format.
-
If the
additional_datasets parameter is omitted from the spec object (the default), then the primary dataset will be used to evaluate each base node in each parse tree.
In this example, we have a constraint string (Equation \ref{dpstr}), which contains two unique base nodes: $\text{(PR | [M])}$ and $\text{(PR | [F])}$. Each base node will get the same additional dataset. The code a few blocks below demonstrates how the additional_datasets dictionary is supplied for this example.
To build our spec object, we first perform the imports and load the primary and additional datasets from the files produced in the Dataset preparation section. The way we load the additional dataset is identical to how we load the primary dataset.
from seldonian.parse_tree.parse_tree import make_parse_trees_from_constraints
from seldonian.dataset import DataSetLoader
from seldonian.utils.io_utils import save_pickle
from seldonian.models.models import BinaryLogisticRegressionModel
from seldonian.models import objectives
if __name__ == '__main__':
primary_cand_data_pth = "gpa_classification_primary_cand_dataset.csv"
primary_safety_data_pth = "gpa_classification_primary_safety_dataset.csv"
primary_metadata_pth = "primary_metadata_classification.json"
addl_cand_data_pth = "gpa_classification_addl_cand_dataset.csv"
addl_safety_data_pth = "gpa_classification_addl_safety_dataset.csv"
addl_metadata_pth = "addl_metadata_classification.json"
regime = "supervised_learning"
sub_regime = "classification"
# Load datasets from file
loader = DataSetLoader(regime=regime)
primary_cand_dataset = loader.load_supervised_dataset(
filename=primary_cand_data_pth,
metadata_filename=primary_metadata_pth,
file_type="csv"
)
primary_safety_dataset = loader.load_supervised_dataset(
filename=primary_safety_data_pth,
metadata_filename=primary_metadata_pth,
file_type="csv"
)
addl_cand_dataset = loader.load_supervised_dataset(
filename=addl_cand_data_pth,
metadata_filename=addl_metadata_pth,
file_type="csv"
)
addl_safety_dataset = loader.load_supervised_dataset(
filename=addl_safety_data_pth,
metadata_filename=addl_metadata_pth,
file_type="csv"
)
The next step is to create the parse tree from the behavioral constraint, demographic parity in this case. We use a for loop to demonstrate how this code generalizes to multiple constraints, though we only have one constraint in this example.
# Behavioral constraints
constraint_strs = ['abs((PR | [M]) - (PR | [F])) - 0.2']
deltas = [0.05]
# For each constraint, make a parse tree
parse_trees = []
for ii in range(len(constraint_strs)):
constraint_str = constraint_strs[ii]
delta = deltas[ii]
# Create parse tree object
parse_tree = ParseTree(
delta=delta,
regime="supervised_learning",
sub_regime=addl_cand_dataset.meta.sub_regime,
columns=addl_cand_dataset.sensitive_col_names,
)
parse_tree.build_tree(constraint_str=constraint_str)
parse_trees.append(parse_tree)
We now create the additional datasets dictionary mapping constraints and their base nodes to the additional candidate and safety dataset objects that we created above.
additional_datasets = {}
for pt in parse_trees:
additional_datasets[pt.constraint_str] = {}
base_nodes_this_tree = list(pt.base_node_dict.keys())
for bn in base_nodes_this_tree:
additional_datasets[pt.constraint_str][bn] = {
"candidate_dataset": addl_cand_dataset,
"safety_dataset": addl_safety_dataset
}
Once filled out, this additional_datasets dictionary will look like this:
{
'abs((PR | [M]) - (PR | [F])) - 0.2': {
'PR | [M]': {
'candidate_dataset': <seldonian.dataset.SupervisedDataSet object at 0x14d083bb0>, 'safety_dataset': <seldonian.dataset.SupervisedDataSet object at 0x14d083ee0>
},
'PR | [F]': {
'candidate_dataset': <seldonian.dataset.SupervisedDataSet object at 0x14d083bb0>, 'safety_dataset': <seldonian.dataset.SupervisedDataSet object at 0x14d083ee0>
}
}
}
Notice in the code block above that we use pt.constraint_str as the outermost key to the additional_datasets dictionary. It is best practice to use this instead of the constraint string we supplied when building the parse tree, for the reasons described above. In our case our constraint string 'abs((PR | [M]) - (PR | [F])) <= 0.2' gets reformatted to 'abs((PR | [M]) - (PR | [F])) - 0.2' when the tree is built.
We also recently introduced the candidate_dataset and safety_dataset parameters to the Spec class. These enable providing the candidate and safety datasets directly, rather than having the engine split the dataset provided in the dataset parameter using the frac_data_in_safety parameter. These two new parameters must be seldonian.dataset.DataSet objects, no different from the requirements of the dataset parameter. If these parameters are not None (the default), then the dataset parameter is ignored. In the code block below, we demonstrate how to provide the primary candidate and safety datasets. First, we setup the remaining objects needed to build the spec object, all of which are the same as those in Tutorial E.
# Model, primary objective
model = BinaryLogisticRegressionModel()
primary_objective = objectives.binary_logistic_loss
frac_data_in_safety = 0.6 # will not be used because we are providing candidate and safety datasets explicitly.
def initial_solution_fn(m,X,y):
return m.fit(X,y)
spec = SupervisedSpec(
dataset=None,
candidate_dataset=primary_cand_dataset,
safety_dataset=primary_safety_dataset,
additional_datasets=additional_datasets,
model=model,
parse_trees=parse_trees,
sub_regime=sub_regime,
frac_data_in_safety=0.6,
primary_objective=primary_objective,
initial_solution_fn=initial_solution_fn,
use_builtin_primary_gradient_fn=True,
optimization_technique='gradient_descent',
optimizer='adam',
optimization_hyperparams={
'lambda_init' : np.array([0.5]),
'alpha_theta' : 0.01,
'alpha_lamb' : 0.01,
'beta_velocity' : 0.9,
'beta_rmsprop' : 0.95,
'use_batches' : False,
'num_iters' : 1000,
'gradient_library': "autograd",
'hyper_search' : None,
'verbose' : True,
}
)
savename = "demographic_parity_addl_datasets_nodups.pkl"
save_pickle(savename,spec,verbose=True)
Below we provide a complete script with all of the steps combined for convenience.
#createspec_nodups.py
from seldonian.parse_tree.parse_tree import *
from seldonian.dataset import DataSetLoader
from seldonian.utils.io_utils import save_pickle
from seldonian.models.models import BinaryLogisticRegressionModel
from seldonian.models import objectives
from seldonian.spec import SupervisedSpec
def initial_solution_fn(m,X,y):
return m.fit(X,y)
if __name__ == '__main__':
primary_cand_data_pth = "gpa_classification_primary_cand_dataset.csv"
primary_safety_data_pth = "gpa_classification_primary_safety_dataset.csv"
primary_metadata_pth = "primary_metadata_classification.json"
addl_cand_data_pth = "gpa_classification_addl_cand_dataset.csv"
addl_safety_data_pth = "gpa_classification_addl_safety_dataset.csv"
addl_metadata_pth = "addl_metadata_classification.json"
regime = "supervised_learning"
sub_regime = "classification"
# Load datasets from file
loader = DataSetLoader(regime=regime)
primary_cand_dataset = loader.load_supervised_dataset(
filename=primary_cand_data_pth,
metadata_filename=primary_metadata_pth,
file_type="csv"
)
primary_safety_dataset = loader.load_supervised_dataset(
filename=primary_safety_data_pth,
metadata_filename=primary_metadata_pth,
file_type="csv"
)
addl_cand_dataset = loader.load_supervised_dataset(
filename=addl_cand_data_pth,
metadata_filename=addl_metadata_pth,
file_type="csv"
)
addl_safety_dataset = loader.load_supervised_dataset(
filename=addl_safety_data_pth,
metadata_filename=addl_metadata_pth,
file_type="csv"
)
# Model, primary objective
model = BinaryLogisticRegressionModel()
primary_objective = objectives.binary_logistic_loss
# Behavioral constraints
constraint_strs = ['abs((PR | [M]) - (PR | [F])) - 0.2']
deltas = [0.05]
# For each constraint, make a parse tree
parse_trees = []
for ii in range(len(constraint_strs)):
constraint_str = constraint_strs[ii]
delta = deltas[ii]
# Create parse tree object
parse_tree = ParseTree(
delta=delta,
regime="supervised_learning",
sub_regime=addl_cand_dataset.meta.sub_regime,
columns=addl_cand_dataset.sensitive_col_names,
)
parse_tree.build_tree(constraint_str=constraint_str)
parse_trees.append(parse_tree)
additional_datasets = {}
for pt in parse_trees:
additional_datasets[pt.constraint_str] = {}
base_nodes_this_tree = list(pt.base_node_dict.keys())
for bn in base_nodes_this_tree:
additional_datasets[pt.constraint_str][bn] = {
"candidate_dataset": addl_cand_dataset,
"safety_dataset": addl_safety_dataset
}
# Save spec object, using defaults where necessary
spec = SupervisedSpec(
dataset=None,
candidate_dataset=primary_cand_dataset,
safety_dataset=primary_safety_dataset,
additional_datasets=additional_datasets,
model=model,
parse_trees=parse_trees,
sub_regime=sub_regime,
frac_data_in_safety=0.6,
primary_objective=primary_objective,
initial_solution_fn=initial_solution_fn,
use_builtin_primary_gradient_fn=True,
optimization_technique='gradient_descent',
optimizer='adam',
optimization_hyperparams={
'lambda_init' : np.array([0.5]),
'alpha_theta' : 0.01,
'alpha_lamb' : 0.01,
'beta_velocity' : 0.9,
'beta_rmsprop' : 0.95,
'use_batches' : False,
'num_iters' : 1000,
'gradient_library': "autograd",
'hyper_search' : None,
'verbose' : True,
}
)
savename = "demographic_parity_addl_datasets_nodups.pkl"
save_pickle(savename,spec,verbose=True)
Running the above Python script will create a pickle file containing the spec object. We can load this pickle file and then use it to run the Seldonian Engine as we normally would. The code below demonstrates how to do this.
This will print out the progress of the optimization process, which should take only about 10 seconds to complete. It will also write out a log file containing the candidate selection details that can be plotted, if desired. We cover how to interpret that plot in other tutorials, such as Tutorial D: Fairness for Automated Loan Approval Systems. In the next section, we cover how to run a Seldonian Experiment when you have additional datasets for the base nodes.
Running a Seldonian Experiment with Additional Datasets
As we saw in the previous section, running the engine with additional datasets requires very little modification beyond the normal procedure. The only requirement is the inclusion of the additional_datasets parameter. We also introduced the candidate_dataset and safety_dataset parameters, but those are not necessary for all additional dataset applications. When running Seldonian Experiments with additional datasets for the base noddes, there are some additional considerations beyond what we have covered so far.
Both the primary dataset and the additional datasets are resampled when the trial datasets are made. After running an experiment with primary and additional datasets, you will notice that there are multiple sets of files in the resampled_datasets/ folder. The resampled primary datasets keep the extant file pattern: resampled_datasets/trial_{trial_index}.pkl, and the additional resampled dataset dictionaries are saved with the pattern resampled_datasets/trial_{trial_index}_addl_datasets.pkl. As with the resampled primary datasets, the resampled additional datasets are of the same size as the original resampled datasets provided in the spec object. This happens behind the scenes, and does not require any user input to enable.
When the spec object contains explicit candidate and safety datasets, as in our example spec object above, the primary resampled datasets are saved with the file pattern: resampled_datasets/trial_{trial_index}_candidate_dataset.pkl and resampled_datasets/trial_{trial_index}_safety_dataset.pkl. The additional resampled dataset files are unaffected.
Seldonian experiments require held-out ground truth datasets to evaluate the first plot (performance) and last plot (probability of constraint violation plot). Until the introduction of additional datasets, both plots were evaluated using a single held out dataset. With the introduction of additional datasets, we added support for providing held out datasets for the primary dataset and additional datasets separately. The held out primary dataset is used to evaluate the performance plot and the held out additional datasets are used to evaluate the probability of constraint violation plot.
The held out primary dataset is provided using the extant pattern, i.e., via the perf_eval_kwargs dictionary parameter to the plot generator object. The held out additional datasets are provided via the "additional_datasets" key to the constraint_eval_kwargs dictionary parameter to the plot generator object. Below is an example script to run a Seldonian experiment using a held out primary dataset and a held out additional dataset for both base nodes. The held out primary dataset is the original dataset from Tutorial E, and the held out additional datasets are the additional datasets we defined for the spec object above. Below is a script to run this experiment. Besides for the modifications just described, the script is very similar to the one used to run the experiment in Tutorial E. We include a binary logistic regressor as a baseline, but exclude the Fairlearn baselines in this tutorial.
import os
os.environ["OMP_NUM_THREADS"] = "1"
import numpy as np
from experiments.generate_plots import SupervisedPlotGenerator
from experiments.baselines.logistic_regression import BinaryLogisticRegressionBaseline
from seldonian.utils.io_utils import load_pickle
from seldonian.dataset import SupervisedDataSet
from sklearn.metrics import log_loss,accuracy_score
def perf_eval_fn(y_pred,y,**kwargs):
# Deterministic accuracy to match Thomas et al. (2019)
return accuracy_score(y,y_pred > 0.5)
def initial_solution_fn(m,X,Y):
return m.fit(X,Y)
def main():
# Parameter setup
run_experiments = True
make_plots = True
save_plot = True
include_legend = False
model_label_dict = {
'qsa':'Seldonian model (with additional datasets)',
}
constraint_name = 'demographic_parity'
performance_metric = 'accuracy'
n_trials = 20
data_fracs = np.logspace(-4,0,15)
n_workers = 8
results_dir = f'results/demographic_parity_nodups'
plot_savename = os.path.join(results_dir,f'gpa_{constraint_name}_{performance_metric}.png')
verbose=False
# Load spec
specfile = f'specfiles/demographic_parity_addl_datasets_nodups.pkl'
spec = load_pickle(specfile)
os.makedirs(results_dir,exist_ok=True)
# Combine primary candidate and safety datasets to be used as ground truth for performance plotd
test_dataset = spec.candidate_dataset + spec.safety_dataset
test_features = test_dataset.features
test_labels = test_dataset.labels
# Setup performance evaluation function and kwargs
perf_eval_kwargs = {
'X':test_features,
'y':test_labels,
'performance_metric':performance_metric
}
# Use original additional_datasets as ground truth (for evaluating safety)
constraint_eval_kwargs = {}
constraint_eval_kwargs["additional_datasets"] = spec.additional_datasets
plot_generator = SupervisedPlotGenerator(
spec=spec,
n_trials=n_trials,
data_fracs=data_fracs,
n_workers=n_workers,
datagen_method='resample',
perf_eval_fn=perf_eval_fn,
constraint_eval_fns=[],
constraint_eval_kwargs=constraint_eval_kwargs,
results_dir=results_dir,
perf_eval_kwargs=perf_eval_kwargs,
)
if run_experiments:
# Logistic regression baseline
lr_baseline = BinaryLogisticRegressionBaseline()
plot_generator.run_baseline_experiment(
baseline_model=lr_baseline,verbose=False)
# Seldonian experiment
plot_generator.run_seldonian_experiment(verbose=verbose)
if make_plots:
plot_generator.make_plots(
tot_data_size=test_dataset.num_datapoints,
fontsize=12,
legend_fontsize=8,
performance_label=performance_metric,
include_legend=include_legend,
model_label_dict=model_label_dict,
save_format="png",
savename=plot_savename if save_plot else None)
if __name__ == "__main__":
main()
Running the script above will produce the following plot (or a very similar one depending on your machine's random number generator):
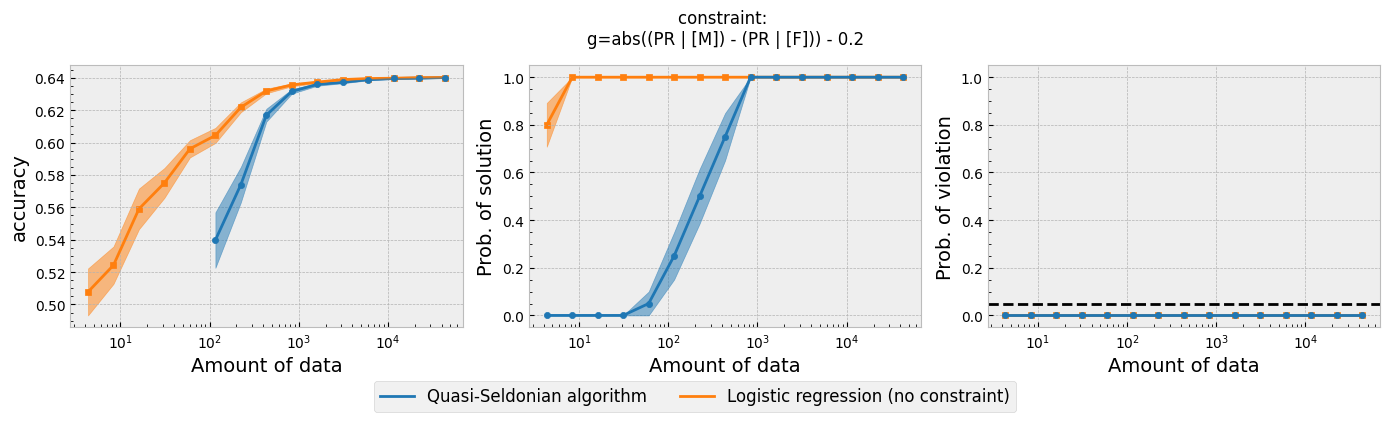 Figure 1 - The three plots of a Seldonian Experiment for the modified GPA classification problem studied in this tutorial.
Figure 1 - The three plots of a Seldonian Experiment for the modified GPA classification problem studied in this tutorial.
The three plots in Figure 1 are similar to the demographic parity plot shown in Figure 1 of Tutorial E, as expected. The main difference is that it appears that more data are needed to achieve a high probability of solution (middle). However, care must be taken when interpreting these three plots when additional datasets are used. The "Amount of Data" label for the horizontal axes refers to the amount of data in the primary dataset for a given data fraction, and gives no indication of the size of additional dataset. One can derive the size of the additional datasets used in any given trial by knowing the total size of the additional datasets and multiplying it by the data fraction. The additional dataset is 30% the size of the full GPA dataset, for reference. For a data fraction of 0.5, for example, the "Amount of Data" would be half the size of the primary dataset, or 43303*0.5 = 21651. For the same data fraction, the size of the additional dataset is 30% of this, or only about 6500. This is why is appears to take more data to achieve the same probability of solution as the experiment in Tutorial E.
Summary
In this tutorial, we demonstrated how to use additional datasets for bounding base nodes. This feature could be useful in scenarios where only a fraction of their data have sensitive attributes. This feature can also be used with completely different datasets for the primary objective and the base nodes. This can be useful when fine-tuning large models subject to behavioral constraints, where one wants to use a pre-existing loss function as the primary objective and custom behavioral constraints.
We demonstrated how to specify additional datasets when building the specification object. We also showed how to use another new feature where the candidate and safety sets are specified explicitly. This feature can be useful on its own (i.e., without using additional datasets), but is particularly helpful when one wants to ensure that there are no shared data points between candidate data and safety data across primary and additional datasets. We first ran the Seldonian Engine using the modified spec object. Then, we ran a Seldonian Experiment, demonstrating how to specify held out datasets when one has additional datasets. We pointed out that interpreting the three plots of a Seldonian Experiment is more complicated when additional datasets are used, as evidenced by the apparent difference betwen the middle plot in Figure 1 compared to the same plot in Tutorial E.